 |
 |
- Search
| Phys Act Nutr > Volume 19(3); 2015 > Article |
|
Abstract
Purpose
We examined whether resistance exercise training restores impaired autophagy functions caused by Chloroquine (CQ)-induced Sporadic Inclusion Body Myositis (sIBM) in rat skeletal muscle.
Methods
Male wistar rats were randomly assigned into three groups: Sham (n = 6), CQ (n = 6), and CQ + Exercise (CE, n = 6). To create a rat model of sIBM, rats in the CQ and CE group were intraperitoneally injected with CQ 5 days a week for 16 weeks. Rats in the CE group performed resistance exercise training 3 times a week for 8 weeks in conjunction with CQ starting from week 9 to week 16. During the training period, maximal carrying load, body weight, muscle weight, and relative muscle weight were measured. Autophagy responses were examined by measuring specific markers.
Results
While maximal carrying capacity for resistance exercise training was dramatically increased in the CE group, no significant changes occurred in the skeletal muscle weight as well as in the relative muscle weight of CE compared to the other groups. CQ treatment caused significant increases in the levels of Beclin-1 and p62, and decreases in the levels of LAMP-2 proteins. Interestingly, no significant differences in the LC3-II/I ratio or the LC3-II protein levels were observed. Although CQ-treatment groups suppressed the levels of the potent autophagy inducer, BNIP3, p62 levels were decreased in only the CE group.
Conclusion
Our findings demonstrate that sIBM induced by CQ treatment results in muscle degeneration via impaired autophagy and that resistance exercise training improves movable loading activity. Finally, regular exercise training may provide protection against sIBM by enhancing the autophagy flux through p62 protein.
Sporadic Inclusion Body Myositis (sIBM) is the most common muscle degenerative disease in elderly populations, with a clinical feature of progressive muscle weakness and severe muscle atrophy [1-3]. The remarkable pathologic features of sIBM present as vacuolated muscle fibers, accumulations of abnormal proteins that are similar to the pathogens of AlzheimerŌĆÖs and ParkinsonŌĆÖs diseases in the brain [4-7]. Although the exact mechanisms of sIBM remain unclear, strong evidence suggests that the primary cause could be endoplasmic reticulum (ER) stress, unfolded protein response, lysosomal inhibition, and dysfunction of protein degradation systems including autophagy [8-10]. Unfortunately, sIBM patients do not respond well to pharmacologic treatments. As a result, exercise has been recommended as a safe method of therapy with no signs of increased muscle inflammation [11-13].
Autophagy is described as a major catabolic process of long-lived bulk proteins and organelles via lysosome-dependent pathways. Following their degradative products, these proteins are recycled during the turnover of intracellular proteins [14]. In all types of autophagy (macro-, micro-, and chaperone-mediated), one prominent feature is the formation of double-membrane vesicles, termed autophagosomes, which eliminate abnormal proteins as well as damaged organelles including mitochondria and transfer them to the lysosome where they are degraded [15]. Basically, autophagy occurs constitutively at lower basal levels in all eukaryotic cells and tissues and is strongly induced by nutrient starvation. However, it is also substantially induced in response to various stressors and signals [16,17]. To activate autophagy, the formation of autophagosomes originating from the control of autophagy-related (Atg) proteins is needed. These Atg proteins modulate autophagy flux, higher rates of initiation and the resolution of autophagic events. The gene profiles emphasize the physiological functions of skeletal muscles in response to exercise [18-21]. Importantly, exercise-induced autophagy is required to maintain skeletal muscle mass and contribute to improving glucose metabolism [22-25]. However, aberrant autophagy flux is detrimental for muscle health and leads to muscle atrophy and degeneration, while sufficient autophagy flux helps maintain healthy myofibers [26]. Accordingly, an optimal level of autophagy is essential in fine-tuning skeletal muscle homeostasis in the physiological state as well as in pathological conditions [27].
Chloroquine (CQ) is a lysosomotropic agent that prevents endosomal acidification. The accumulation of CQ in lysosomes causes an increase in lysosomal pH, inhibiting the activities of lysosomal proteases and eventually diminishing autophagy flux [28]. Furthermore, the long-term effects of CQ treatment in rats cause myopathy involving histological changes as well as the accumulation of excessive autophagosomes and ╬▓-amyloid in their skeletal muscle fibers [29,30]. For that reason, this experimental approach could be a useful tool for better understanding of the molecular mechanisms in muscle degenerative diseases with common pathogenic phenotypes followed by the identification of autophagy signaling pathways in rats as well as in sIBM humans [31-34].
Resistance exercise is an effective method to increase muscle mass and strength [35]. Therefore, resistance training is an important therapeutic strategy that should be considered for the purpose of improving functioning, the ability to perform activities of daily living and health-related quality of life [36]. Recently, wheel running was reported to alleviate the detrimental effects of CQ on skeletal muscles in mice by repairing autophagy flux [37]. However, not only are the effects of long-term resistance training-induced autophagy in muscular disease yet to be fully defined, but the precise cellular function of Atg proteins in the autophagy signaling pathways remain to be explained [38]. Therefore, this is the first study to investigate whether the autophagy signaling pathways will provide a protective mechanism in the skeletal muscles of rats with sIBM induced by CQ treatment following long-term progressive resistance exercise training.
Male Wistar Hannover rats (N = 18, 6 weeks of age) were obtained from the Central Laboratory Animal Incorporation (Seoul, Korea). These rats were housed in standard rodent cages with unrestricted access to water and food (temperature of 22┬░C, humidity 50-60%, ventilation- and light-controlled 12h to 12h light-dark cycle). The body weight of the rats was measured five times a week at the same time. The adaptation period was set for 2 weeks, after which the 8 weeks old rats were randomly assigned into three groups: 1) Control (Sham, n = 6), 2) Chloroquine (CQ, n = 6), 3) Chloroquine + Exercise (CE, n = 6).
The resistance training protocol for muscle disease rat models has not been examined so far. Thus, we aimed to establish a protocol applicable to subjects with muscle disease in our previous test. Referencing existing protocols [39-42], our modified method was accomplished successfully. The workout equipment consisting of climbing ladders (20 ├Ś 115 cm) with 4 cm grids inclined at 85 degrees were made in our laboratory. The rats in the CE group were trained 3 days/week (Monday, Wednesday, and Friday) for 8 weeks, for a total of 24 training sessions. In the first week, the CE group was familiarized with the ladder through voluntary climbing from the bottom to the top of the ladder. After familiarization, the resistance training began using lead weights attached to the base of the tail. To evaluate the 1 RM of CE group rats, a carrying load of 50% of the ratŌĆÖs body weight was attached to each ratŌĆÖs tail and gradually incremented until they reached their maximum carrying load. After determining the 1 RM, the CE group climbed the ladder with 50% (1st-2nd time), 75% (3rd-4th time), and 100% (5th-6th time) of the previous maximal carrying load. The resistance training consisted of 10 repetitions per training session, and when they reached the top of the ladder, they were allowed to recover in the resting area for 2 min. If a rat was able to climb the ladder with these loads, additional weights were placed in 50mL conical tubes at 30 g increments for each subsequent climb.
At the end of the experimental period, all animals in each group were acutely anesthetized with pentobarbital sodium (40 mg/kg i.p). After reaching a surgical plane of anesthesia, soleus muscles were removed, frozen immediately in liquid nitrogen and stored at ŌłÆ80┬░C for subsequent analyses. We assumed that the effects of CQ were not the same for each muscle fiber type. Based upon CQ-induced myopathy studies, muscle atrophy occurred predominantly in type I muscles and unusual autophagosomes were accumulated [43,44]. Therefore, this study focused exclusively on the slow-twitch soleus muscle as the representative skeletal muscle.
For protein analysis, the soleus muscles were homogenized with RIPA buffer (Biosesang, R2002, Korea) containing a Halt Protease and Phosphatase inhibitor cocktail (Thermoscientific, 78446, USA) using a homogenizer (Automill, Tokken Co., Japan). After incubation for 30 min on ice, centrifugation was conducted at 14,000 RCF for 30 min at 4┬░C. The resulting supernatant was collected, and protein content was assessed by the BCA method (Pierce BCA Protein Assay Kit, 23225, USA). For the running step, 40 ╬╝g of each protein was separated by SDS-PAGE with 8~15% gels for 1 hour and 30min at 80 Volts due to the molecular size of the target proteins and the proteins were transferred to PVDF membranes for 1 hour at 100 Volts. Non-specific sites were blocked for 1 hour at room temperature in the blocking solution (5% w/v BSA, 1X TBS, and 0.1% Tween-20) and then the membranes were incubated overnight at 4┬░C with primary antibodies. The following antibodies were used: Beclin-1 (3738, 1:1000), Atg7 (2631, 1:1000), BNIP3 (3769, 1:1000), p62 (5114, 1:1000), and LC3 (4108, 1:1000) from Cell Signaling; LAMP-2 (sc-8100, 1:200) from Santa Cruz Biotechnology; Cathepsin L (ab133641, 1:1000) from abcam. In addition, following incubation with primary antibodies, the membranes were washed extensively with 1X TBS-T for 30 min and then incubated with secondary antibodies HRP-conjugated goat anti-rabbit (1148960, 1:5000), HRP-conjugated rabbit anti-goat (811620, 1:2000) from Invitrogen, and HRP-conjugated goat anti-mouse (SC-2005, 1:5000) from Santa Cruz. After repeating the washing steps three times for 30 min at room temperature, immunoreactivity was detected with luminataTM Forte Western HRP Substrate (WBLUF 0100, Millipore, USA) in accordance with the manufacturerŌĆÖs instructions. The digital images were acquired using a ChemiDoc XRS System (Bio-Rad, USA). Band intensities were quantified by densitometric analysis using Image Lab Software (Bio-Rad, USA) and values were normalized to a-tubulin for the loading control (sc-5286, Santa Cruz Biotechnology).
All data analyses were conducted with SPSS 18.0 software (SPSS, Chicago, IL, USA). The results are presented as means ┬▒ SE. Comparisons between groups for each dependent variable were made by a one-way ANOVA. If statistical significance was found among groups, TukeyŌĆÖs honestly significantly different tests were performed for the post hoc analysis. Significance levels were established at ╬▒< 0.05.
The initial body weights among the groups were not significantly different at the early experimental stage, which established homogeneity among the groups (Table 1). In contrast, the final body weights (Sham: 443.5 ┬▒ 14.9 g, CQ: 376.0 ┬▒ 11.9 g, CE: 341.8 ┬▒ 10.3 g) were significantly different in CQ (p < 0.01) and CE (p < 0.001) compared to Sham. Thus, it was necessary to normalize muscle mass for differences in body weight as relative muscle weight. As a result, we evaluated the soleus muscle weight and the relative weight of the muscle. However, there was no significant difference between soleus muscle weight and relative soleus muscle weight among groups.
All rats in the CE group successfully completed the entire eight weeks of resistance training. The training protocols were measured by their ability to carry progressively heavier loads. After 24 training sessions, the rats in the CE group significantly increased their maximal carrying capacity from baseline to completion by 246% (p < 0.001, Fig. 1).
Over the course of 24 training sessions (3 times/week, total 8 weeks) the ratŌĆÖs maximal carrying capacity increased 246% from baseline (315.0 g ┬▒ 20.5) to the end of the training period (776.0 g ┬▒ 41.7). Results are represented as means ┬▒ SE (n = 6).
We examined the activation of the autophagy signaling pathway that begins with the formation of autophagosomes and involves a series of cascades. As a marker for the initiation of autophagosome formation, Beclin-1 protein levels were significantly increased in CQ compared to Sham (p < 0.05, Fig. 2A-B). However, the amount of elongation and formation markers for the autophagosome Atg7 protein were not significantly different among the groups, although Atg7 expression showed a decreasing tendency in CQ compared to Sham (Fig. 2A, 2C). In addition, as an inducer of mitochondrial autophagy downstream of the transcription factor FoxO3a, BNIP3 protein levels significantly decreased in both CQ (p < 0.01) and CE (p < 0.05) compared to Sham (Fig. 2A, 2D). To investigate the autophagy flux, we measured the protein levels of p62 and LC3-II as well as the LC3-II/I ratio. One of the best characterized factors of selective autophagy, p62 is an adaptor of autophagosome formation and the protein was overexpressed in CQ compared to Sham, while p62 levels in CE were significantly decreased compared to CQ (p < 0.001, Fig. 2A, 2E). Interestingly, LC3-II and the LC3 II/I ratio, which are key markers of autophagy, were not significantly different among the groups (Fig. 2A, 2F-G).
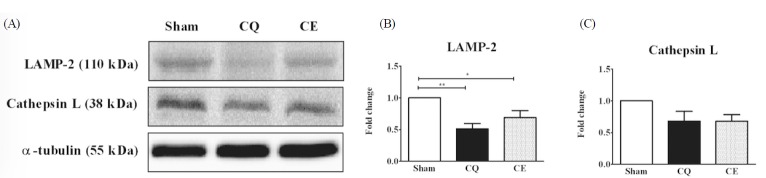
In the final step of autophagy, which involves the fusion of autophagosomes with lysosomes, LAMP-2 (Lysosome-associated membrane protein-2) and Cathepsin L proteins are characterized by an autophagosomal-lysosome degradation system. Specifically, LAMP-2 protein levels were significantly decreased in CQ (p < 0.01) and CE (p < 0.05) groups compared to Sham (Fig. 3A-B). However, Cathepsin L protein levels were not significantly different among the groups (p > 0.05), although the protein seemed to be downregulated in both CQ and CE groups compared to Sham (Fig. 3A, 3C).
This study is the first to investigate whether proper regulation of autophagy signaling-related proteins will provide therapeutic strategies or muscle strength improvement in sIBM-induced animal rat models after 8 weeks of resistance training. In addition, we also investigated whether the rats gradually lost body weight including muscle mass. In fact, most of the studies in vitro have been focused on the pathogenic features for abnormality instead of physical alterations [30,45]. Thus, we measured and identified whether changes in body weight, soleus muscle weight, and relative soleus muscle weight due to muscle atrophy are characterized by clinical phenotype. In our data, the final body weights in the two CQ-treated groups were dramatically decreased (Table 1). However, our experimental values didnŌĆÖt show significant differences in either the soleus muscle weight or the relative muscle weight, despite the expectation of dynamic changes in the groups. Based upon the features of severe muscle atrophy, loss of body weight in CQ-treated rats are supported as the trend for chronological changes [29]. Amazingly, a few studies have reported the accumulation of vacuolated muscle fibers and ╬▓-amyloid concurrent with autophagy activation in the soleus muscles of CQ-treated rats [32,44]. Accordingly, we suspect that the autophagy signaling pathway could contribute to muscle disease development. To confirm this hypothesis, it is necessary to examine the overall steps of autophagy.
Beclin-1 is a pro-apoptotic Bcl-2 homology domain (BH3)-only protein, unlike the members of the anti-apoptotic Bcl-2 family of proteins [46]. The mammalian ortholog of yeast Atg6/Vps30, Beclin-1, is one of the essential Atg proteins that affect autophagy induction and is associated with diverse biological as well as pathological processes [47]. In the present study, Beclin-1 protein levels were significantly increased in CQ (Fig. 2A-B). A recent study demonstrated increased expression of Beclin-1 in muscle biopsies of sIBM patients and the results indicated its function in increasing sequestrating through the double-membrane of autophagosomes as well as endosome formation [34]. Although altered Beclin-1 expression levels have been observed in multiple diseases including neurodegenerative, heart, and cancer, the role of Beclin-1 in skeletal muscle disease has not been explored yet. Thus, our finding demonstrates the expression of Beclin-1 in the skeletal muscles of CQ-induced muscle disease rat models for the first time.
Atg7 is an autophagy-related E1-like enzyme that serves as the elongation and formation marker for autophagosomes and is essential for the E2-substrate reaction of LC3-lipidation [48]. Specifically, muscle-specific deletion of the Atg7 gene resulted in severe muscle atrophy and dysfunction [26]. In our results, Atg7 protein levels were not significantly different among the groups (Fig. 2A, 2C), even though the Atg7 levels showed a decrease in both CQ and CE groups compared to Sham. In fact, it is still unexplored to the Atg7 protein in sIBM patients or CQ-treated myopathy animal rats. Therefore, we suggest that it is necessary to investigate the function of Atg7 in a variety of conditions that could lead to muscle dysfunction.
BNIP3 is a pro-apoptotic BH3 only protein that induces mitochondrial dysfunction and cell death. BNIP3 is also a potent inducer of autophagy [49]. Our results revealed that BNIP3 protein levels were significantly decreased in both CQ and CE compared to Sham (Fig. 2A, 2D). Contrary to these results, the BNIP3 level was significantly increased in CQ-treated female ICR mice [37].
The autophagy flux is a very important index that explains the rates of initiation and resolution of autophagic events for maintaining muscle mass and myofiber integrity [25]. Accordingly, we monitored the expression of p62 and LC3-II proteins as well as the LC3-II/I ratio. The p62 protein, also known as sequestosome 1 (SQSTM 1), is a shuttle protein that facilitates the delivery of polyubiquitinated protein aggregates and is a common component exposed in protein aggregation diseases through interactions with LC3 at the autophagosome [50,51]. Therefore, the accumulation of p62 protein is associated with autophagy suppression, while proper turnover of p62 regulates autophagy by preventing spontaneously formed aggregates [52]. Our data showed that the p62 levels of CE significantly decreased compared to CQ (p < 0.001), while the p62 protein levels significantly increased in CQ compared to Sham (Fig. 2A, 2E). The overexpression of p62 in the CQ group in our study corresponds with findings in sIBM muscle fibers [6]. Up-regulated p62 protein expression in CQ was observed in female ICR mice, while p62 level was down-regulated in the exercise group [37]. This study showed that wheel running exercise improved deficient autophagy activation by eliminating the detrimental effect of CQ on skeletal muscles in mice. Therefore, we suggest that progressive long-term resistance exercise training could be used as a therapeutic strategy in myopathy with muscle atrophy to improve muscle function and strength.
LC3, the mammalian homolog of yeast Atg8, is localized in autophagosome membranes and has been used as an autophagosome marker to monitor autophagy. LC3 is also easily integrated into intracellular protein aggregates [53]. Two forms of endogenous LC3 are observed, the unconjugated form of LC3-I and the conjugated form of LC3-II, followed by the conversion (LC3-I to LC3-II) of LC3 when autophagy is induced [54]. To identify the autophagy activity, we detected the amount of LC3-II, which is associated with the number of autophagosomes and degraded by lysosomal hydrolases [55]. Interestingly, our data showed that the protein levels of LC3-II and the LC3 II/I ratio were not significantly different among the groups (Fig. 2A, 2E-F). In contrast, other studies reported the overexpression of LC3-II following experimental CQ-injection in rat muscles [31,32] as well as in sIBM human muscles [34]. We attributed our results to a ceiling effect because of the long-term CQ treatment. However, the results may also be due to defective autophagy resulting from long term CQ-treatment, with an excess amount of LC3-II [56].
LAMP-2 (Lysosome associated membrane protein-2) is a critical protein that is required for fusion of the autophagosome with the lysosome [57,58]. A few studies showed that increased expression of LAMP-2 in myopathies with rimmed vacuole accumulation results in abnormal lysosome function [59-61]. Contradicting these studies, our results showed that the expression of LAMP-2 proteins decreased in both CQ-treated groups compared to Sham (Fig. 3A-B). Supporting our results, other studies have also reported that LAMP-2 deficiency can be a potent inducer of myopathies evidenced in human DanonŌĆÖs disease and LAMP-2 deficient mice [62,63]. For these reasons, LAMP-2 protein is proposed to have alternative roles in various disease conditions as well as species differences. Therefore, we investigated selective disturbances in lysosomal functions from the decrease in LAMP-2 levels caused by long-term CQ-treatment.
Cathepsin L is a lysosomal protease that is present in the lysosome. It is identified as a skeletal muscle atrophy marker, playing a key role in protein turnover via lysosomal degradation corresponding to the final steps of the autophagy signaling pathways [64]. Recent investigations report that the lysosomal enzyme activities of cathepsin D and B in sIBM muscles were decreased in a manner similar to our results (Fig. 3A, 3C), whereas the protein expression levels were increased [10]. Moreover, there was evidence that muscle atrophy in calpain-3 deficient mice and cathepsin L mRNA levels decreased in the skeletal muscles [65].
Our findings demonstrate that sIBM induced by CQ treatment results in muscle degeneration via impaired autophagy and that long-term resistance exercise training promotes autophagy, thus attenuating sIBM-induced muscle degeneration caused by autophagy dysfunction. Finally, our data indicate that regular resistance training provides potential protection against sIBM by enhancing autophagy flux through the p62 protein.
ACKNOWLEDGMENT
This work was supported by the National Research Foundation of Korea Grant funded by the Korean Government (NRF-2013S1A5A8023992). All processes for experiments on living animals were approved by the Institutional Animal Care and Use Committee at Korea National Sport University (Certificate KNSU-IACUC-2013-01).
REFERENCES
1. Needham M, Corbett A, Day T, Christiansen F, Fabian V, Mastaglia FL. Prevalence of sporadic inclusion body myositis and factors contributing to delayed diagnosis. Journal of clinical neuroscience : official journal of the Neurosurgical Society of Australasia 2008;15:1350-3. PMID: 18815046.


2. Cox FM, Titulaer MJ, Sont JK, Wintzen AR, Verschuuren JJ, Badrising UA. A 12-year follow-up in sporadic inclusion body myositis: an end stage with major disabilities. Brain : a journal of neurology 2011;134:3167-75. PMID: 21908393.



3. Cortese A, Machado P, Morrow J, Dewar L, Hiscock A, Miller A, Brady S, Hilton-Jones D, Parton M, Hanna MG. Longitudinal observational study of sporadic inclusion body myositis: implications for clinical trials. Neuromuscular disorders : NMD 2013 23:404-12. Epub 2013/03/16. PMID: 23489664.


4. Askanas V, Engel WK, Nogalska A. Pathogenic considerations in sporadic inclusion-body myositis, a degenerative muscle disease associated with aging and abnormalities of myoproteostasis. Journal of neuropathology and experimental neurology 2012 71:680-93. Epub 2012/07/19. PMID: 22805774.


5. Vattemi G, Nogalska A, King Engel W, DŌĆÖAgostino C, Checler F, Askanas V. Amyloid-beta42 is preferentially accumulated in muscle fibers of patients with sporadic inclusion-body myositis. Acta neuropathologica 2009;117:569-74. PMID: 19280202.



6. Nogalska A, Terracciano C, DŌĆÖAgostino C, King Engel W, Askanas V. p62/SQSTM1 is overexpressed and prominently accumulated in inclusions of sporadic inclusion-body myositis muscle fibers, and can help differentiating it from polymyositis and dermatomyositis. Acta neuropathologica 2009;118:407-13. PMID: 19557423.



7. Askanas V, Engel WK. Sporadic inclusion-body myositis: conformational multifactorial ageing-related degenerative muscle disease associated with proteasomal and lysosomal inhibition, endoplasmic reticulum stress, and accumulation of amyloid-beta42 oligomers and phosphorylated tau. Presse medicale (Paris, France : 1983) 2011 40:e219-35. Epub 2011/03/12.


8. Vattemi G, Engel WK, McFerrin J, Askanas V. Endoplasmic reticulum stress and unfolded protein response in inclusion body myositis muscle. Am J Pathol 2004;164:1-7. PMID: 14695312.



9. Askanas V, Engel WK, Nogalska A. Inclusion body myositis: a degenerative muscle disease associated with intra-muscle fiber multi-protein aggregates, proteasome inhibition, endoplasmic reticulum stress and decreased lysosomal degradation. Brain pathology 2009;19:493-506. PMID: 19563541.



10. Nogalska A, DŌĆÖAgostino C, Terracciano C, Engel WK, Askanas V. Impaired autophagy in sporadic inclusion-body myositis and in endoplasmic reticulum stress-provoked cultured human muscle fibers. Am J Pathol 2010;177:1377-87. PMID: 20616343.



11. Alexanderson H. Exercise in inflammatory myopathies, including inclusion body myositis. Current rheumatology reports 2012 14:244-51. Epub 2012/04/03. PMID: 22467380.



12. Gualano B, Neves M Jr, Lima FR, Pinto AL, Laurentino G, Borges C, Baptista L, Artioli GG, Aoki MS, Moriscot A, Lancha AH Jr, Bonfa E, Ugrinowitsch C. Resistance training with vascular occlusion in inclusion body myositis: a case study. Medicine and science in sports and exercise 2010;42:250-4. PMID: 19927034.


13. Spector SA, Lemmer JT, Koffman BM, Fleisher TA, Feuerstein IM, Hurley BF, Dalakas MC. Safety and efficacy of strength training in patients with sporadic inclusion body myositis. Muscle & nerve 1997 20:1242-8. Epub 1997/10/27 20:24. PMID: 9324080.


14. Rajawat YS, Hilioti Z, Bossis I. Aging: central role for autophagy and the lysosomal degradative system. Ageing research reviews 2009 8:199-213. Epub 2009/05/12. PMID: 19427410.


15. Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. The Journal of pathology 2010;221:3-12. PMID: 20225336.


16. Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Molecular biology of the cell 2004;15:1101-11. PMID: 14699058.



17. Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nature reviews Molecular cell biology 2009;10:458-67. PMID: 19491929.



18. Kim YA, Kim YS, Song W. Autophagic response to a single bout of moderate exercise in murine skeletal muscle. Journal of physiology and biochemistry 2012;68:229-35. PMID: 22205581.



19. Kim YA, Kim YS, Oh SL, Kim HJ, Song W. Autophagic response to exercise training in skeletal muscle with age. Journal of physiology and biochemistry 2013;69:697-705. PMID: 23471597.



20. Lira VA, Okutsu M, Zhang M, Greene NP, Laker RC, Breen DS, Hoehn KL, Yan Z. Autophagy is required for exercise training-induced skeletal muscle adaptation and improvement of physical performance. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 2013;27:4184-93. PMID: 23825228.



21. Bayod S, Del Valle J, Pelegri C, Vilaplana J, Canudas AM, Camins A, Jimenez A, Sanchez-Roige S, Lalanza JF, Escorihuela RM, Pallas M. Macroautophagic process was differentially modulated by long-term moderate exercise in rat brain and peripheral tissues. Journal of physiology and pharmacology : an official journal of the Polish Physiological Society 2014 65:229-39. Epub 2014/05/02. PMID: 24781732.

22. Grumati P, Coletto L, Schiavinato A, Castagnaro S, Bertaggia E, Sandri M, Bonaldo P. Physical exercise stimulates autophagy in normal skeletal muscles but is detrimental for collagen VI-deficient muscles. Autophagy 2011;7:1415-23. PMID: 22024752.



23. Nair U, Klionsky DJ. Activation of autophagy is required for muscle homeostasis during physical exercise. Autophagy 2011 7:1405-6. Epub 2011/11/16. PMID: 22082869.



24. He C, Bassik MC, Moresi V, Sun K, Wei Y, Zou Z, An Z, Loh J, Fisher J, Sun Q, Korsmeyer S, Packer M, May HI, Hill JA, Virgin HW, Gilpin C, Xiao G, Bassel-Duby R, Scherer PE, Levine B. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature 2012;481:511-5. PMID: 22258505.




25. Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M, Metzger D, Reggiani C, Schiaffino S, Sandri M. Autophagy is required to maintain muscle mass. Cell metabolism 2009;10:507-15. PMID: 19945408.


26. Masiero E, Sandri M. Autophagy inhibition induces atrophy and myopathy in adult skeletal muscles. Autophagy 2010 6:307-9. Epub 2010/01/28. PMID: 20104028.


27. Sandri M. Autophagy in skeletal muscle. FEBS letters 2010 584:1411-6. Epub 2010/02/06. PMID: 20132819.


28. Kumamoto T, Ueyama H, Watanabe S, Murakami T, Araki S. Effect of denervation on overdevelopment of chloroquine-induced autophagic vacuoles in skeletal muscles. Muscle & nerve 1993 16:819-26. Epub 1993/08/01. PMID: 8332133.


29. Sugita H, Higuchi I, Sano M, Ishiura S. Trial of a cysteine proteinase inhibitor, EST, in experimental chloroquine myopathy in rats. Muscle & nerve 1987 10:516-23. Epub 1987/07/01. PMID: 3306368.


30. Tsuzuki K, Fukatsu R, Takamaru Y, Kimura K, Abe M, Shima K, Fujii N, Takahata N. Immunohistochemical evidence for amyloid beta in rat soleus muscle in chloroquine-induced myopathy. Neuroscience letters 1994 182:151-4. Epub 1994/12/05. PMID: 7715800.


31. Kimura N, Kumamoto T, Kawamura Y, Himeno T, Nakamura KI, Ueyama H, Arakawa R. Expression of autophagy-associated genes in skeletal muscle: an experimental model of chloroquine-induced myopathy. Pathobiology : journal of immunopathology, molecular and cellular biology 2007;74:169-76.


32. Ikezoe K, Furuya H, Arahata H, Nakagawa M, Tateishi T, Fujii N, Kira J. Amyloid-beta accumulation caused by chloroquine injections precedes ER stress and autophagosome formation in rat skeletal muscle. Acta neuropathologica 2009;117:575-82. PMID: 19198858.



33. Lunemann JD, Schmidt J, Schmid D, Barthel K, Wrede A, Dalakas MC, Munz C. Beta-amyloid is a substrate of autophagy in sporadic inclusion body myositis. Annals of neurology 2007 61:476-83. Epub 2007/05/01. PMID: 17469125.


34. Girolamo F, Lia A, Amati A, Strippoli M, Coppola C, Virgintino D, Roncali L, Toscano A, Serlenga L, Trojano M. Overexpression of autophagic proteins in the skeletal muscle of sporadic inclusion body myositis. Neuropathology and applied neurobiology 2013;39:736-49. PMID: 23452291.


35. Murton AJ, Greenhaff PL. Resistance exercise and the mechanisms of muscle mass regulation in humans: acute effects on muscle protein turnover and the gaps in our understanding of chronic resistance exercise training adaptation. Int J Biochem Cell Biol 2013;45:2209-14. PMID: 23872221.


36. Alexanderson H, Lundberg IE. Exercise as a therapeutic modality in patients with idiopathic inflammatory myopathies. Current opinion in rheumatology 2012;24:201-7. PMID: 22189517.


37. Jiang D, Chen K, Lu X, Gao HJ, Qin ZH, Lin F. Exercise ameliorates the detrimental effect of chloroquine on skeletal muscles in mice via restoring autophagy flux. Acta pharmacologica Sinica 2014;35:135-42. PMID: 24335841.



38. Tam BT, Siu PM. Autophagic cellular responses to physical exercise in skeletal muscle. Sports medicine (Auckland, NZ) 2014 44:625-40. Epub 2014/02/20.


39. Begue G, Douillard A, Galbes O, Rossano B, Vernus B, Candau R, Py G. Early activation of rat skeletal muscle IL-6/STAT1/STAT3 dependent gene expression in resistance exercise linked to hypertrophy. PloS one 2013;8:e57141PMID: 23451164.



40. Hornberger TA Jr, Farrar RP. Physiological hypertrophy of the FHL muscle following 8 weeks of progressive resistance exercise in the rat. Canadian journal of applied physiology=Revue canadienne de physiologie appliquee 2004 29:16-31. Epub 2004/03/06. PMID: 15001801.


41. Lee S, Barton ER, Sweeney HL, Farrar RP. Viral expression of insulin-like growth factor-I enhances muscle hypertrophy in resistance-trained rats. Journal of applied physiology 2004;96:1097-104. PMID: 14766764.


42. Luo L, Lu AM, Wang Y, Hong A, Chen Y, Hu J, Li X, Qin ZH. Chronic resistance training activates autophagy and reduces apoptosis of muscle cells by modulating IGF-1 and its receptors, Akt/mTOR and Akt/FOXO3a signaling in aged rats. Experimental gerontology 2013 48:427-36. Epub 2013/02/20. PMID: 23419688.


43. Schmalbruch H. The early changes in experimental myopathy induced by chloroquine and chlorphentermine. Journal of neuropathology and experimental neurology 1980 39:65-81. Epub 1980/01/01. PMID: 7359173.



44. Suzuki T, Nakagawa M, Yoshikawa A, Sasagawa N, Yoshimori T, Ohsumi Y, Nishino I, Ishiura S, Nonaka I. The first molecular evidence that autophagy relates rimmed vacuole formation in chloroquine myopathy. Journal of biochemistry 2002 131:647-51. Epub 2002/05/02. PMID: 11983070.



45. Tsuzuki K, Fukatsu R, Takamaru Y, Yoshida T, Hayashi Y, Yamaguchi H, Fujii N, Takahata N. Amyloid beta protein in rat soleus muscle in chloroquine-induced myopathy using end-specific antibodies for A beta 40 and A beta 42: immunohistochemical evidence for amyloid beta protein. Neuroscience letters 1995 202:77-80. Epub 1995/12/29. PMID: 8787835.


46. Cao Y, Klionsky DJ. Physiological functions of Atg6/Beclin 1: a unique autophagy-related protein. Cell research 2007;17:839-49. PMID: 17893711.



47. He C, Levine B. The Beclin 1 interactome. Current opinion in cell biology 2010;22:140-9. PMID: 20097051.



48. Tanida I, Yamasaki M, Komatsu M, Ueno T. The FAP motif within human ATG7, an autophagy-related E1-like enzyme, is essential for the E2-substrate reaction of LC3 lipidation. Autophagy 2012;8:88-97. PMID: 22170151.


49. Zhang J, Ney PA. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell death and differentiation 2009 16:939-46. Epub 2009/02/21. PMID: 19229244.




50. Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. The Journal of cell biology 2005;171:603-14. PMID: 16286508.




51. Zatloukal K, Stumptner C, Fuchsbichler A, Heid H, Schnoelzer M, Kenner L, Kleinert R, Prinz M, Aguzzi A, Denk H. p62 Is a Common Component of Cytoplasmic Inclusions in Protein Aggregation Diseases. The American Journal of Pathology 2002;160:255-63. PMID: 11786419.



52. Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, Hamazaki J, Nishito Y, Iemura S, Natsume T, Yanagawa T, Uwayama J, Warabi E, Yoshida H, Ishii T, Kobayashi A, Yamamoto M, Yue Z, Uchiyama Y, Kominami E, Tanaka K. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 2007 131:1149-63. Epub 2007/12/18. PMID: 18083104.


53. Kuma A, Matsui M, Mizushima N. LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy 2007 3:323-8. Epub 2007/03/28. PMID: 17387262.


54. Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. The EMBO journal 2000 19:5720-8. Epub 2000/11/04. PMID: 11060023.



55. Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annual review of nutrition 2007;27:19-40.


56. Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy 2007 3:542-5. Epub 2007/07/06. PMID: 17611390.


57. Eskelinen EL, Illert AL, Tanaka Y, Schwarzmann G, Blanz J, Von Figura K, Saftig P. Role of LAMP-2 in lysosome biogenesis and autophagy. Molecular biology of the cell 2002 13:3355-68. Epub 2002/09/11. PMID: 12221139.



58. Huynh KK, Eskelinen EL, Scott CC, Malevanets A, Saftig P, Grinstein S. LAMP proteins are required for fusion of lysosomes with phagosomes. The EMBO journal 2007 26:313-24. Epub 2007/01/25. PMID: 17245426.



59. Tsuruta Y, Furuta A, Furuta K, Yamada T, Kira J, Iwaki T. Expression of the lysosome-associated membrane proteins in myopathies with rimmed vacuoles. Acta neuropathologica 2001 101:579-84. Epub 2001/08/23. PMID: 11515786.



60. Kumamoto T, Ueyama H, Tsumura H, Toyoshima I, Tsuda T. Expression of lysosome-related proteins and genes in the skeletal muscles of inclusion body myositis. Acta neuropathologica 2004;107:59-65. PMID: 14513262.



61. Cacciottolo M, Nogalska A, DŌĆÖAgostino C, Engel WK, Askanas V. Chaperone-mediated autophagy components are upregulated in sporadic inclusion-body myositis muscle fibres. Neuropathology and applied neurobiology 2013;39:750-61. PMID: 23452232.


62. Nishino I, Fu J, Tanji K, Yamada T, Shimojo S, Koori T, Mora M, Riggs JE, Oh SJ, Koga Y, Sue CM, Yamamoto A, Murakami N, Shanske S, Byrne E, Bonilla E, Nonaka I, DiMauro S, Hirano M. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease). Nature 2000 406:906-10. Epub 2000/09/06. PMID: 10972294.



63. Tanaka Y, Guhde G, Suter A, Eskelinen EL, Hartmann D, Lullmann-Rauch R, Janssen PM, Blanz J, von Figura K, Saftig P. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature 2000 406:902-6. Epub 2000/09/06. PMID: 10972293.



64. Deval C, Mordier S, Obled C, Bechet D, Combaret L, Attaix D, Ferrara M. Identification of cathepsin L as a differentially expressed message associated with skeletal muscle wasting. The Biochemical journal 2001 360:143-50. Epub 2001/11/07. PMID: 11696001.




65. Combaret L, Bechet D, Claustre A, Taillandier D, Richard I, Attaix D. Down-regulation of genes in the lysosomal and ubiquitin-proteasome proteolytic pathways in calpain-3-deficient muscle. Int J Biochem Cell Biol 2003;35:676-84. PMID: 12672459.


Fig.┬Ā2
Protein levels of Beclin-1, Atg7, BNIP3, p62, LC3-II and LC3-II/I ratio in soleus muscles were analyzed as an essential marker involved in processes for the autophagosome formation. A) Representative western blot images of Beclin-1, Atg7, BNIP3, p62, LC3-II, LC3 II/I ratio and ╬▒-tubulin among groups. B) Quantification of Beclin-1 protein. C) Quantification of Atg7 protein. D) Quantification of BNIP3 protein. E) Quantification of p62 protein. F) Quantification of LC3-II protein. G) Quantification of LC3-II/I protein ratio. Protein expression comparisons were performed after normalization to ╬▒-tubulin. Results are represented as means ┬▒ SE (n = 6 rats/group). * p < 0.05, ** p < 0.01, *** p < 0.001 vs. Sham; #
p < 0.001 vs. CE.
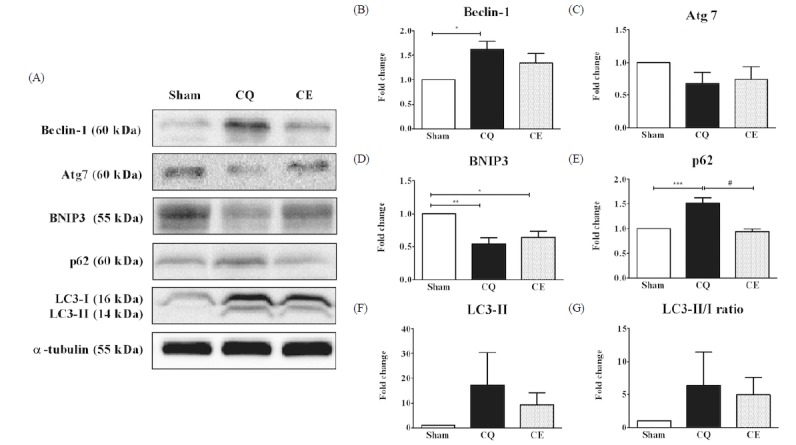
Fig.┬Ā3
Protein levels of LAMP-2 and Cathepsin L in soleus muscles were analyzed as a receptor at the lysosomal membrane and a marker of proteolytic capacity of lysosome, respectively. A) Representative western blot images of LAMP-2 and Cathepsin L among groups. Protein expression comparisons were performed after normalization to ╬▒-tubulin. B) Quantification of LAMP-2 protein. C) Quantification of Cathepsin L protein. Results are represented as means ┬▒ SE (n = 6 rats/group). * p < 0.05, ** p < 0.01 vs. Sham
Table┬Ā1
Comparisons of body weight, muscle weight, and relative muscle weight among three groups during 18 weeks
| Sham (n = 6) | CQ (n = 6) | CE (n = 6) | p | |
|---|---|---|---|---|
| Initial body weight (g) | 236.7 ┬▒ 4.4 | 232.3 ┬▒ 4.2 | 243.5 ┬▒ 4.3 | 0.215 |
| Final body weight (g) | 443.5 ┬▒ 14.9 | 376.0 ┬▒ 11.9** | 341.8 ┬▒ 10.3*** | < .001 |
| Soleus (mg) | 243.3 ┬▒ 9.5 | 210.0 ┬▒ 10.0 | 210.0 ┬▒ 14.4 | 0.093 |
| Soleus/body weight (mg) | 55.1 ┬▒ 2.3 | 55.9 ┬▒ 2.3 | 61.3 ┬▒ 3.3 | 0.237 |
-
METRICS

-
- 17 Crossref
- 1,998 View
- 28 Download
- Related articles in Phys Act Nutr






